Hadepot: Repository of MapReduce Applications
The goal of this repository is to give researchers easy access to existing, publicly available MapReduce applications. These applications can serve as examples for writing new applications. They can also serve as benchmark or simply sample workloads for new systems, tools, and techniques for performing large-scale, complex analytics in the cloud.
The repository is curated:
| University of Washington | Duke University |
|---|---|
|
YongChul Kwon Magdalena Balazinska Bill Howe |
Herodotos Herodotou Rozemary Scarlat Shivnath Babu |
How to contribute?
To propose a new application for inclusion in the repository, please send as much of the following information as possible via email to hadepot@cs.washington.edu.
- Title and a short description of the application
- A link to the code
- A link to the dataset
- A job history file generated by running code on data
- Any performance challenges encountered with this application (exhibits skew, long-running, failure-sensitive, etc.).
- If possible, also include any relevant documentation to help get the application set up and running.
List of Applications
Click to see the detail of each application.
PageRank
Description: PageRank is a link analysis algorithm that assigns weights (ranks) to each vertex in a graph by iteratively aggregating the weights of its inbound neighbors.
- Application Areas: web, graph analysis
- Author: Jimmy Lin
- Source: [Cloud 9 Documentation] [Github]
- License: Apache License 2.0
- Dataset: Billion Triple Challenge 2010, Freebase data dump
- Hadoop Compatibility: 0.20 or higher. Old API.
- Characteristics:
For basic implementation, a map task becomes expensive when the task encounters a node having extremely high out degree (root cause of a few slow tasks in the figure below)
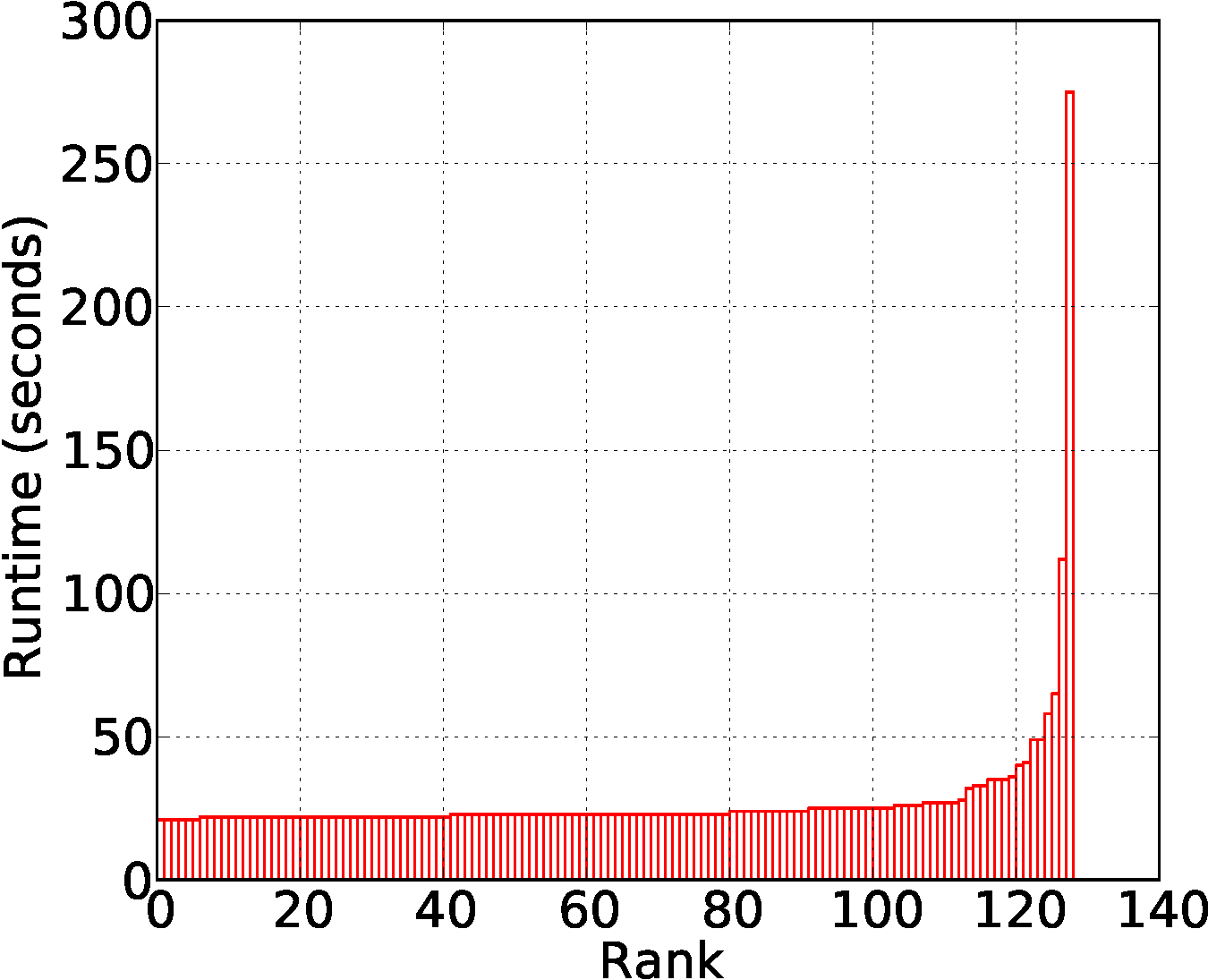
- Submitted By: yongchul
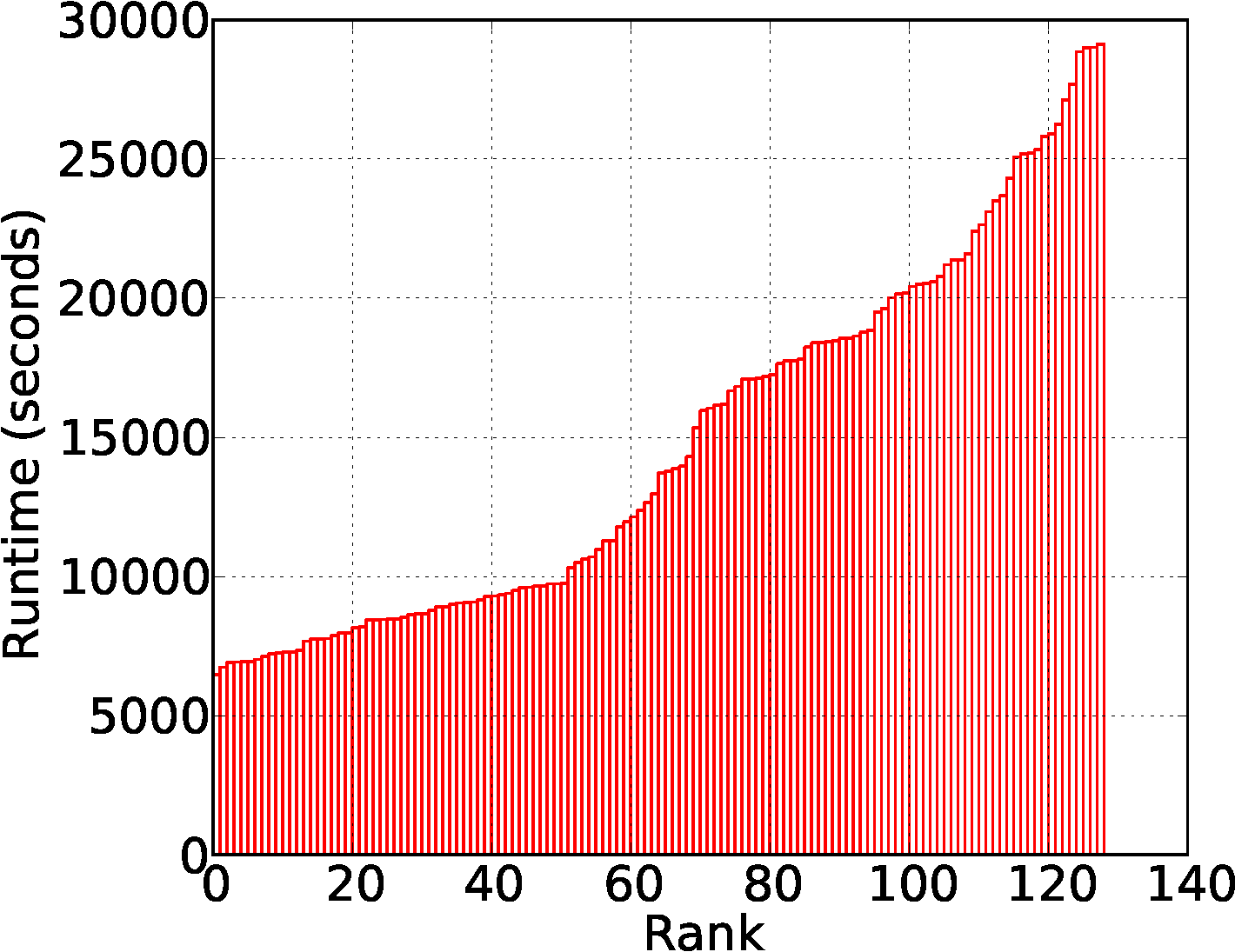
CloudBurst
Description: CloudBurst is a MapReduce implementation of the RMAP algorithm for short-read gene alignment2. CloudBurst aligns a set of genome sequence reads with a reference sequence. CloudBurst distributes the approximate-alignment computations across reduce tasks by partitioning the reads and references on their n-grams.
- Application Areas: bioinformatics, short-read sequence analysis
- Author: Michael Schatz
- Source: http://cloudburst-bio.sourceforge.net/
- Hadoop Compatibility: 0.20 or higher. Old API.
- Characteristics:
For map, the runtime distribution is a bi-modal distribution of which modes correspond to the two input datasets.
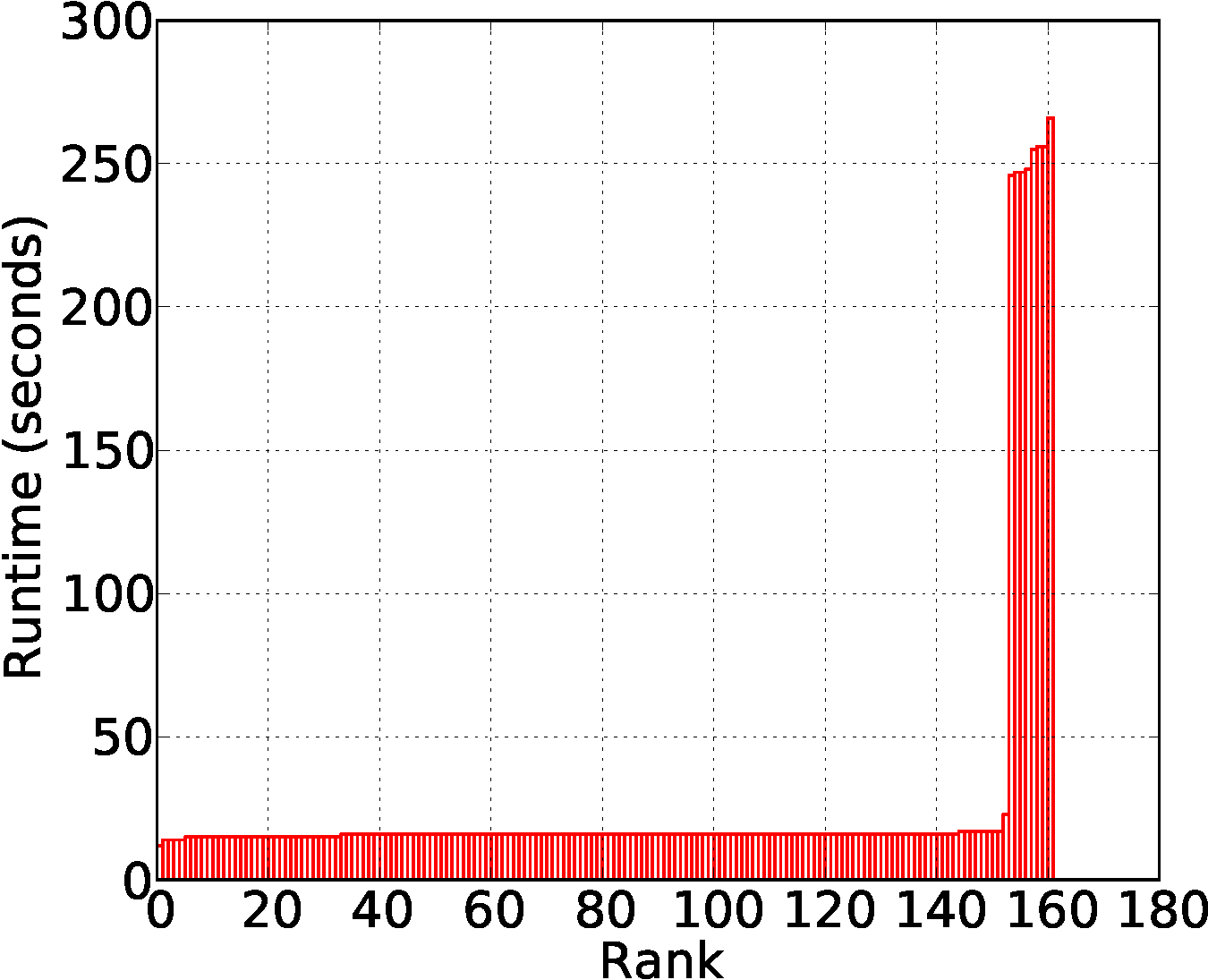
Reduce is computationally intensive. The runtime distribution of reduce tasks is relatively uniform.
Distributed Friends-of-Friends
Description: FoF is used by astronomers to analyze the structure of the universe within a snapshot of a simulation of the universe evolution. For each point in the dataset, the FoF algorithm uses a spatial index to recursively look up neighboring points to find connected clusters.
- Application Areas: Astronomy, astrophysics simulations
- Author: YongChul Kwon
- Source:
- License: Apache License 2.0
- Dataset: Please contact yongchul
- Hadoop Compatibility: 0.20 or higher. New API.
- Characteristics:
The runtime of local clustring phase (map) is sensitive to the distribution of input data in 3D space.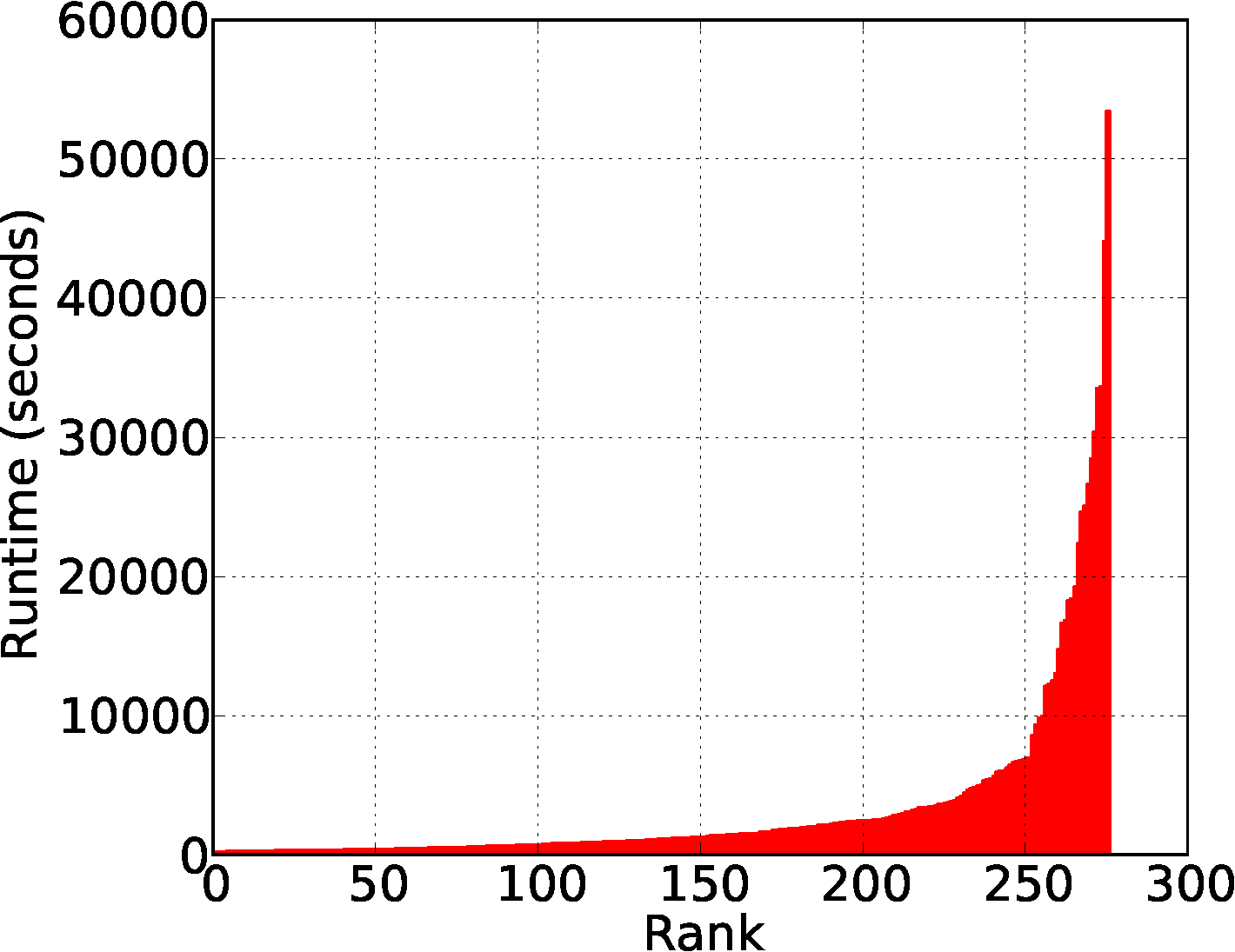
- Submitted By: yongchul
MR-Tandem: Parallel Peptide Search with X!Tandem and Hadoop MapReduce
Description: MR-Tandem adapts the popular X!Tandem peptide search engine to work with Hadoop MapReduce for reliable parallel execution of large searches. MR-Tandem supports local Hadoop clusters as well as Amazon Web Services for creating inexpensive on-demand Hadoop clusters, enabling search volumes that might not otherwise be feasible with the compute resources a researcher has at hand. MR-Tandem is designed to drop in wherever X!Tandem is already in use and requires no modification to existing X!Tandem parameter files or workflows. The work of moving data into the cluster and search results back onto the local file system is handled transparently.
- Application Areas: Bioinformatics, Proteomics, Peptide Search
- Author: Brian Pratt, Insilicos LLC (extends X!Tandem work by Beavis et al)
- Source:
- License: Apache License 2.0
- Dataset: Many search sets are available at http://www.peptideatlas.org/repository/
- Hadoop Compatibility: 0.18 or higher. Hadoop Streaming.
- Characteristics:
Excellent scalability improvement over existing brittle MPI-based parallel peptide search efforts, but parallelism is still limited by the need for each search task to create the same list of theoretical spectra. - Submitted By: Brian Pratt (www.insilicos.com)
Hadoop MapReduce Example
Description: This package contains a subset of the Hadoop 0.20.2 example MapReduce programs adapted to use the new MapReduce API.
- Application Areas: General
- Author: The Hadoop team, Herodotos Herodotou
- Source: Download
- License: Apache License 2.0
- Dataset: Data generators are included in the source code above.
- Hadoop Compatibility: 0.20.2 or higher. New API.
- Submitted: Herodotos Herodotou
PigMix Benchmark
Description: PigMix is a set of queries used test pig performance from release to release. There are queries that test latency and queries that test scalability. In addition it includes a set of map reduce java programs to run equivalent map reduce jobs directly. Original code taken from https://issues.apache.org/jira/browse/PIG-200 and modified by Herodotos Herodotou to simplify usage and use Hadoop's new API. See enclosed README file for detailed instructions on how to generate the data and execute the benchmark. You will find further details at https://cwiki.apache.org/confluence/display/PIG/PigMix
- Application Areas: General, Benchmark
- Author: Daniel Dai, Alan Gates, Amir Youseffi, Herodotos Herodotou
- Source: Download
- License: Apache License 2.0
- Dataset: The data generator is included in the source code above and can be executed using the
pigmix/scripts/generate_data.shscript. - Hadoop Compatibility: 0.20.2 or higher. New API. Pig.
- Submitted By: Herodotos Herodotou
Hive Performance Benchmark
Description: This benchmark was taken from https://issues.apache.org/jira/browse/HIVE-396 and was slight modified to make it easier to use. The queries appear in the Pavle et. al. paper A Comparison of Approaches to Large-Scale Data Analysis. See enclosed README file for detailed instructions on how to generate the data and execute the benchmark.
- Application Areas: General, Benchmark
- Author: Yuntao Jia, Zheng Shao
- Source: Download
- License: Apache License 2.0
- Dataset: The data generator is included in the source code above in the
datagendirectory. - Hadoop Compatibility: 0.20.2 or higher. New API. Hive. Pig.
- Submitted By: Herodotos Herodotou
TPC-H Benchmark
Description: TPC-H is an ad-hoc, decision support benchmark (http://www.tpc.org/tpch/). The attached source ode contains (i) scripts for generating TPC-H data in parallel and loading them on HDFS, (ii) the Pig-Latin implementation of all TPC-H queries, and (iii) the HiveQL implementation of all TPC-H queries (see https://issues.apache.org/jira/browse/HIVE-600).
- Application Areas: General, Benchmark
- Author: Herodotos Herodotou, Jie Li, Yuntao Jia
- Source: Download
- License: Apache License 2.0
- Dataset: See enclosed README file for detailed instructions on how to generate the data and execute the benchmark
- Hadoop Compatibility: 0.20.2 or higher. Hive.
- Submitted By: Herodotos Herodotou
Term Frequency-Inverse Document Frequency
Description:
The TF-IDF weight (Term Frequency-Inverse Document Frequency) is a weight often used in information retrieval and text mining. This weight is a statistical measure used to evaluate how important a word is to a document in a collection or corpus. For details, please visit: http://en.wikipedia.org/wiki/Tf%E2%80%93idf.
See enclosed README file for detailed instructions on how to execute the jobs. TF-IDF is implemented as a pipeline of 3 MapReduce jobs:
- Calculates word frequency in documents
- Calculates word counts for documents
- Counts the documents in the corpus and computes the TF-IDF
- Application Areas: Information Retrieval, Text, Document
- Author: Herodotos Herodotou, (code adapted from Marcello de Sales)
- Source: Download
- License: Apache License 2.0
- Dataset: Any set of text documents
- Hadoop Compatibility: 0.20.2 or higher. New API.
- Submitted By: Herodotos Herodotou
Behavioral Targeting (Model Generation)
Description:
This application builds a model that can be used to score ads for each user based on its search and click behavior. The model consists of a weight assigned to each (ad, keyword) pair and that shows the correlation between the two (meaning that a larger weight signifies a greater probability that a user will click on an ad if he has searched for that keyword). The job takes as input the path to the folder that contains 3 log files named clickLog, impressionsLog and searchLog. The impressionsLog records when an ad (identified by the adId) was shown to a user (identified by the userId). The clickLog records when a user clicked on an ad. The searchLog records when a user searched for a particular keyword. These values are assumed to be comma separated and the order of the fields is: timestamp, userId, adId (or keyword in the searchLog).
See enclosed BT_description.pdf file for detailed instructions on how to execute the jobs.
- Application Areas: Web Search, Data Mining, Machine Learning, Ad, Log Analysis
- Author: Rozemary Scarlat
- Source: Download
- License: Apache License 2.0
- Dataset: The data generator is included in the source code above.
- Hadoop Compatibility: 0.20.2 or higher. New API.
- Submitted By: Herodotos Herodotou
Various Pig Workflows
Description:
This package includes pig scripts and dataset generators for various pig workflows.
See enclosed README file for detailed instructions on how to execute the pig scripts. The Pig-Latin scripts included are:
coauthor.pig: Finds the top 20 coauthorship pairs. The input data has "paperid \t authorid" records. It first creates a list of authorids for each paperid. Then, it count the author pairs. Then it finds the author pairs with the highest count.pagerank.pig: Runs 1 iteration of page rank. The input datasets are Rankings ("pageid \t rank") and Pages ("pageid \t comma-delimited links").pavlo.pig: Complex join task from the Sigmod 08 Pavlo et. al. paper.tf-idf.pig: Computes TF-IDF. The input data has "doc \t word" records.tpch-17.pig: Query 17 from the TPC-H benchmark but slightly modified to have a different orderings of columns (this script assumes that part key is the first column of both tables).common.pig: A seven job workflow. The first job scans and performs an initial processing of the data. Two jobs read, filter, and find the sum and maximum of the prices for each {order ID, part ID}, {orderID, supplier ID}, respectively. The results of these two jobs are further processed by separate jobs to find the overall sum and maximum prices for each {order ID}. Finally, the results are separately used to find the number of distinct aggregated prices.
- Application Areas: General, Benchmark
- Author: Harold Lim
- Source: Download
- License: Apache License 2.0
- Dataset: The data generator is included in the source code above.
- Hadoop Compatibility: 0.20.2 or higher. Pig.
- Submitted By: Herodotos Herodotou



